Structure/Function Studies of Transcription Factor Drivers in Cancer
Our lab is fundamentally interested in understanding how transcription factors that are drivers in cancer mediate their effects. This basic science understanding is essential to develop new approaches to cancer treatment. Our approach to do this is based on structure/function studies. We determine 3D structures of functional domains from these transcription factors that mediate specific protein-protein or protein-nucleic acid interactions bound to their interaction partner. Based on the structural data, we develop specific point mutations which disrupt this specific interaction but do not affect the structure or stability of the domain. These mutant forms of the transcription factor serve as high quality biological reagents to carry out functional studies with. Furthermore, they recapitulate what a small molecule inhibitor of the interaction would do, so they serve as tools to validate specific interactions for inhibitor development. Using these well-validated biological tools, we probe functional effects including, but not limited to, effects on proliferation, effects on differentiation (flow cytometry), effects on gene expression (RNA-Seq), effects on transcription factor occupancy (ChIP-Seq), effects on epigenetic signaling (ChIP-Seq), and effects in relevant mouse models of the cancer (latter is done with a group of outstanding collaborators at other institutions).
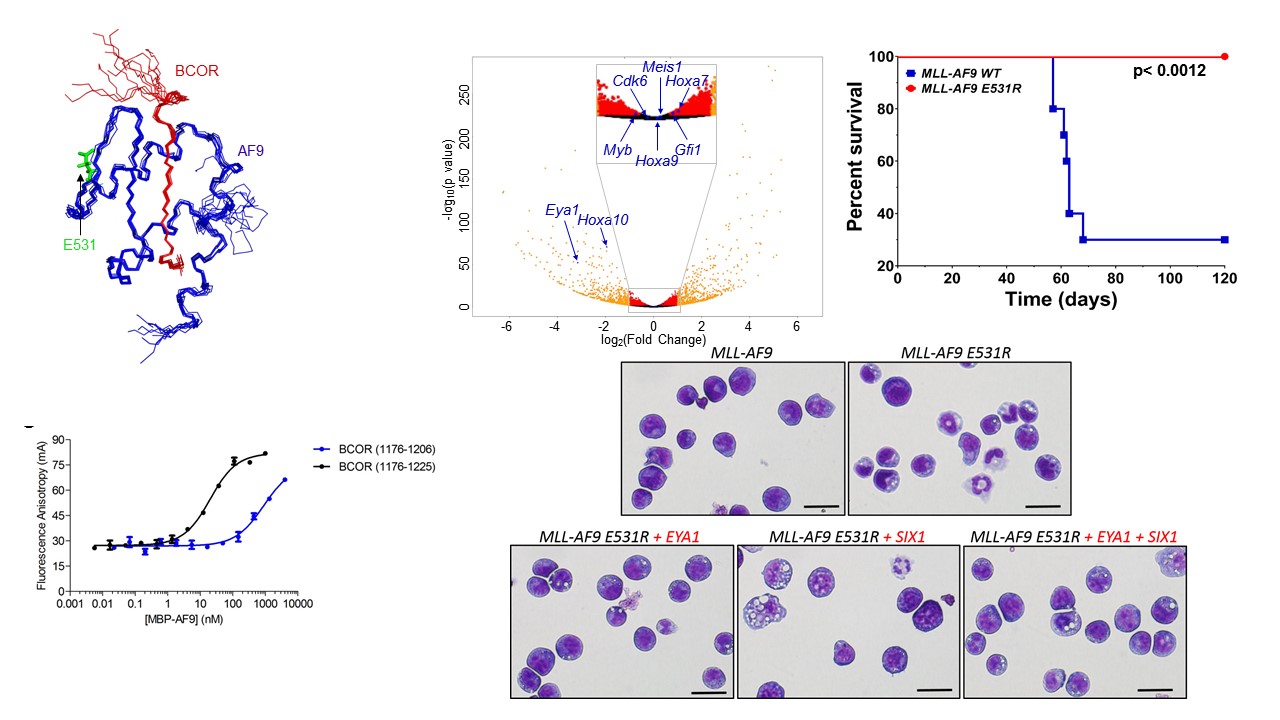
A long-term focus has been structural studies of a novel transcription factor referred to as the core-binding factor (CBF) which is a heterodimeric transcription factor (CBFβ and RUNX1, 2, or 3). CBFβ/RUNX1 is essential for hematopoietic development. Gene translocations associated with the genes coding for CBFβ and RUNX1 produce novel fusion proteins (CBFβ-SMMHC, RUNX1-ETO, TEL-RUNX1) which have been implicated as playing a role in more than 30% of acute leukemias. We have carried out extensive structural and functional studies of the fusion protein forms of CBFβ and RUNX1. We have extended these studies to the MLL protein, a key epigenetic regulator that is the target of chromosomal translocations (MLL-AF9, MLL-ENL, MLL-AF4, etc.) in leukemia which are particularly poor prognosis.
Example publications:
- Cierpicki T, Risner LE, Grembecka J, Lukasik SM, Popovic R, Omonkowska M, Shultis DD, Zeleznik-Le NJ, Bushweller JH. Structure of the MLL CXXC domain-DNA complex and its functional role in MLL-AF9 leukemia. Nat Struct Mol Biol. 2010 Jan;17(1):62-8. PubMed PMID: 20010842; PubMed Central PMCID: PMC2908503.
- Kuntimaddi A, Achille NJ, Thorpe J, Lokken AA, Singh R, Hemenway CS, Adli M, Zeleznik-Le NJ, Bushweller JH. Degree of recruitment of DOT1L to MLL-AF9 defines level of H3K79 Di- and tri-methylation on target genes and transformation potential. Cell Rep. 2015 May 5;11(5):808-20. PubMed PMID: 25921540; PubMed Central PMCID: PMC4426023.